

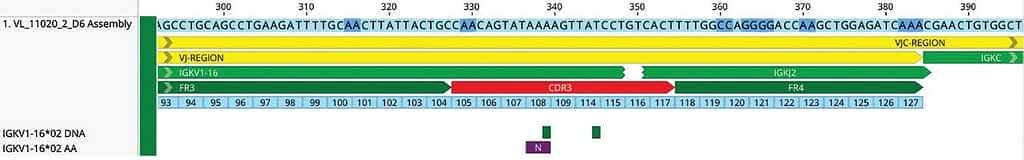
a clear difference between one sequence to the others (Remember this can be possible if the sequences are distantly related but still cross reference the alignment to the individual assemblies).
GENEIOUS PRIME REMOVE GAPS DOWNLOAD
You may download the fasta file of both, however, the alignment of amino acids is what will be used for the second quality check.
GENEIOUS PRIME REMOVE GAPS CODE
In the Genetic Code box select the relevant reading frame and be sure to check the “Guess most likely reading frame” option. We suggest you leave the Protein Alignment Option method selected as “Muscle”.
GENEIOUS PRIME REMOVE GAPS PDF
You find al the commands and an executable example in the Sample_Data! Help Center with context-sensitive help and tutorials PTC Mathcad Prime 5.0.0.0 Read This First in PDF format To access the Help Center or the Getting Started Tutorial, click or press F1. You can also click any item on the user interface Ribbon or any function in the worksheet and press F1 to open the relevant Help topic. Generate the configuration file by runing batch_config("config.txt")in R. The QuikChange® Primer Design Program supports mutagenic primer design for your QuikChange mutagenesis experiments. #GENEIOUS TUTORIAL PRIMER DESIGN DOWNLOAD#Įdit this file to review and change the default settings.Ĭreate a csv table containing the groups (and their subgroups if you want to download a subset of that group) for which data should be downloaded. Using primer design guidelines described in QuikChange manuals, this program calculates/designs the appropriate primer sequences with the optimal melting temperature.

See "taxa_small.csv" in the folder Sample_Data. PrimerMiner is a R based batch sequence downloader to design and verify metabarcoding primers. To start the batch download, run batch_download("taxa_small.csv", "config.txt") giving the name of the taxon table and configuration file.Īlign OTUs and extract region interesting for primer development e.g. Export the aligned sequences as a fasta file. You can apply selective trimming on the edges to remove primer sequences from the alignment as well as remove gaps from the alignemnt with the selectivetrim() function. With plot_alignments(path_to_fasta_alignment_files) you can produce plots of the alignments, to use for primer design and visualisation (see Poster_introduction.pdf for an example).


 0 kommentar(er)
0 kommentar(er)
